A Kinetic Approach to Determining the Conformational Stability of a Protein That Dimerizes after Folding

By A Mystery Man Writer

A) Typical kinetic unfolding curves of GFP-cycle3 at pH 6.2 caused by

Conformational stability and folding mechanisms of dimeric proteins - ScienceDirect

Molecules, Free Full-Text

Protein kinetic stability - ScienceDirect

Dimer Formation of a Stabilized Gβ1 Variant: A Structural and

PDF) Measuring the Conformational Stability of a Protein by Hydrogen Exchange

Thermodynamic stability of proteins

Protein conformational plasticity and complex ligand-binding kinetics explored by atomistic simulations and Markov models
Molecules, Free Full-Text

Franz X. Schmid's research works Johannes Gutenberg-Universität

Protein electrostatics: From computational and structural analysis to discovery of functional fingerprints and biotechnological design - Computational and Structural Biotechnology Journal

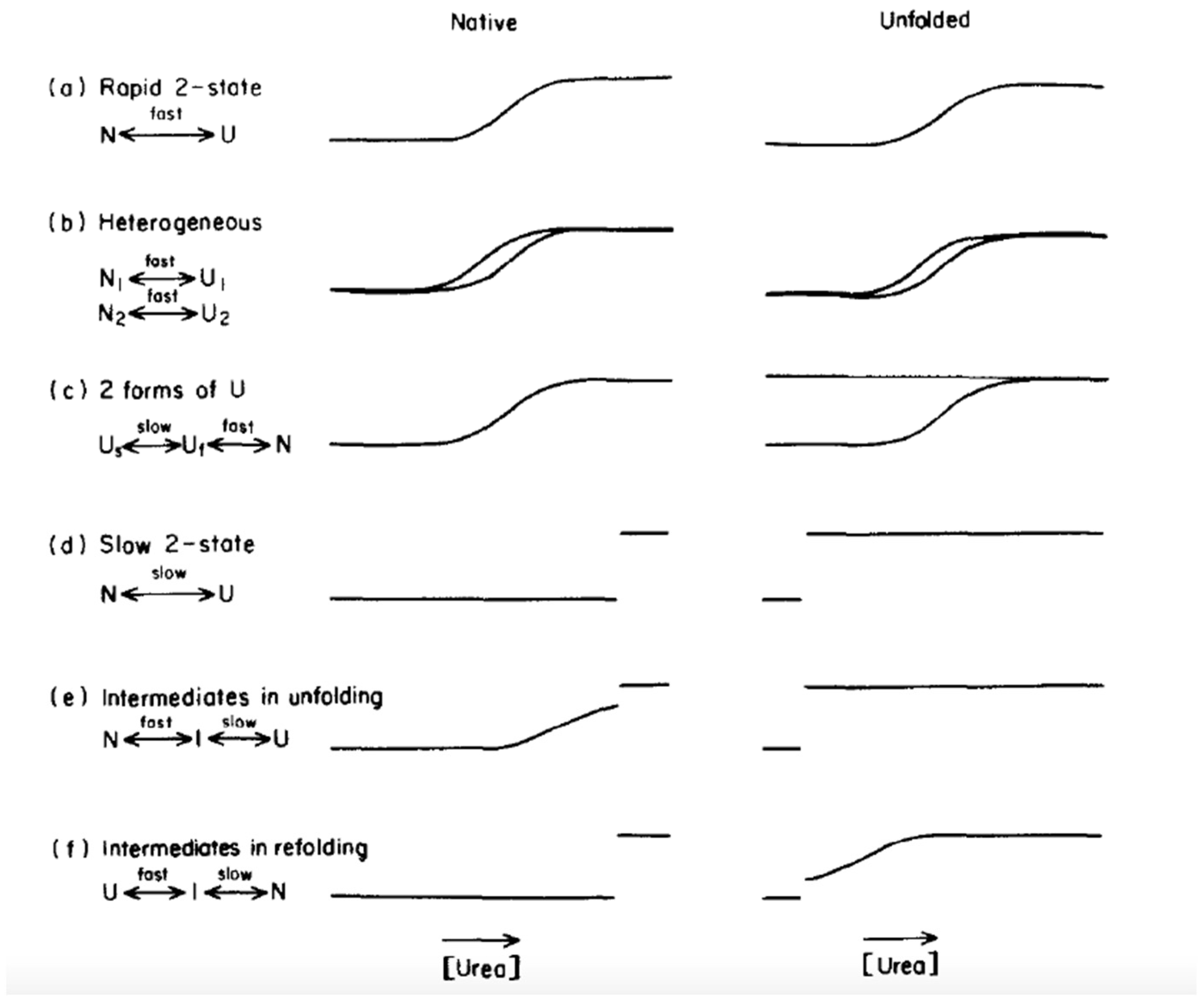
Schematic diagram and free energy profiles for protein folding. (a)
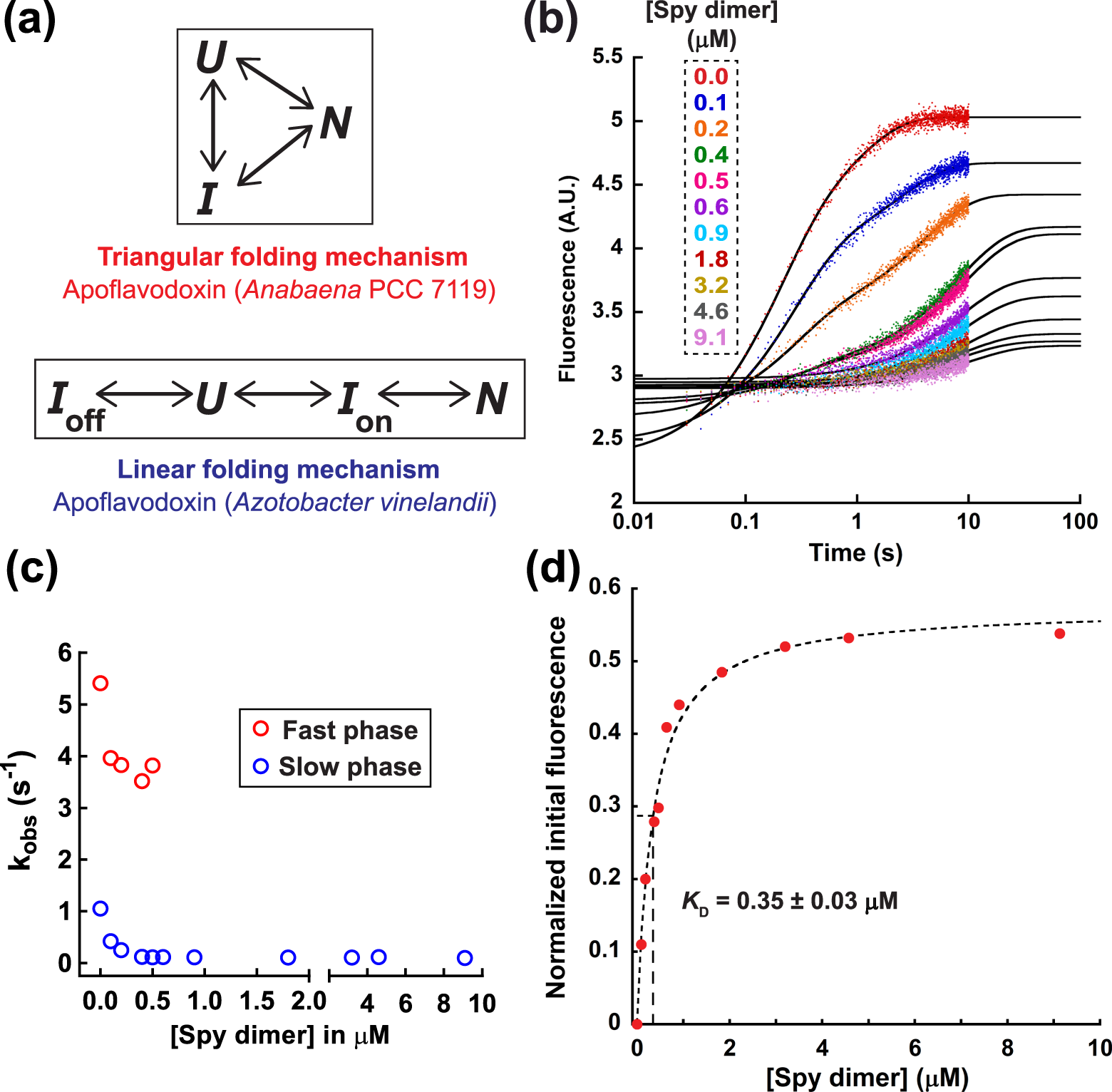
Mechanism of the small ATP-independent chaperone Spy is substrate specific
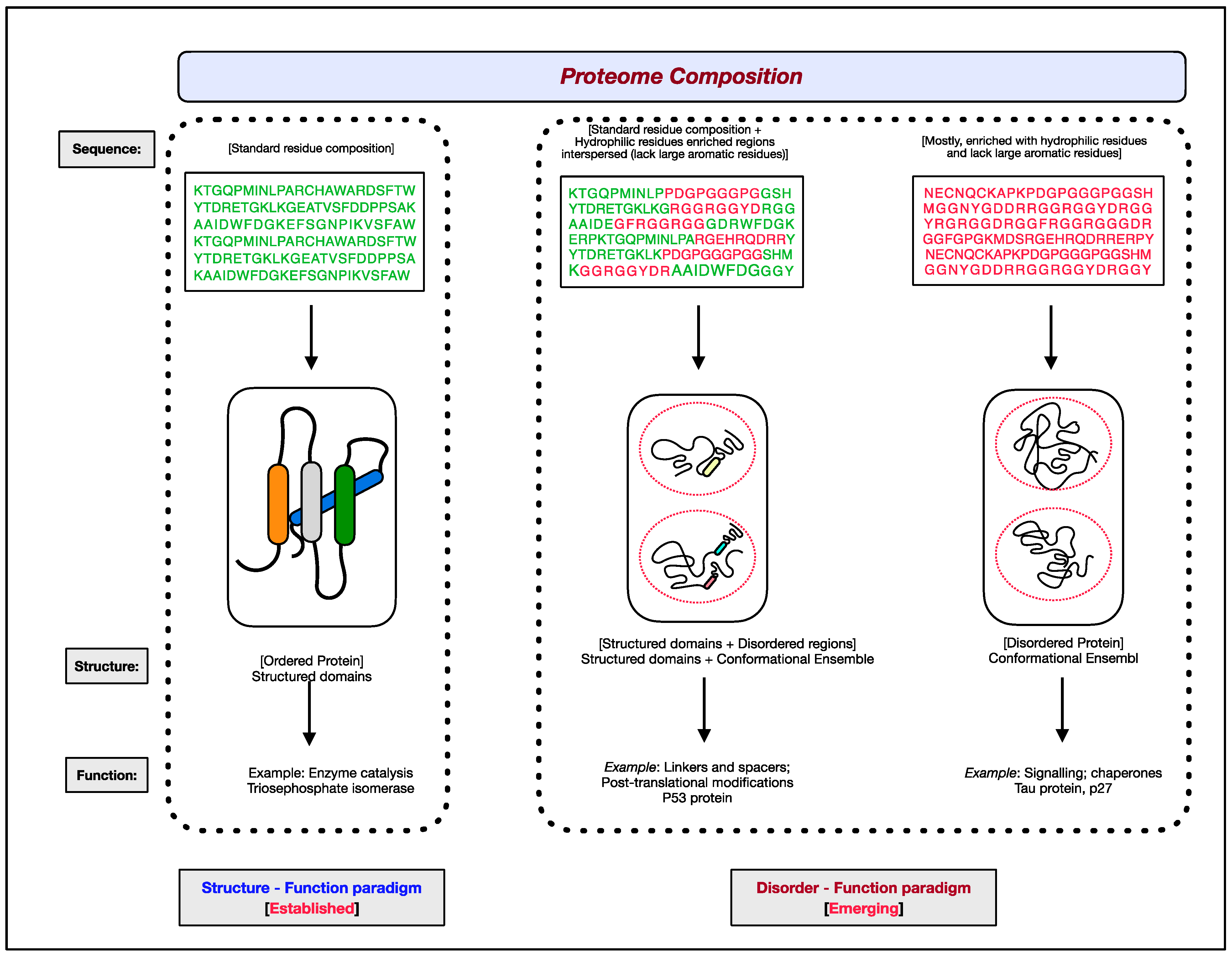
IJMS, Free Full-Text

Conformational stability and folding mechanisms of dimeric proteins - ScienceDirect
- 90 Degree By Reflex Womens Power Flex Yoga Pants Ghana

- Монголын хөрөнгийн зах зээл 30 нас хүрлээ

- Here are Saturday's high school sports results for the Appleton and Green Bay area

- Sale On Yoga Pants International Society of Precision Agriculture

- Flibanserin‐Stimulated Partner Grooming Reflects Brain Metabolism Changes in Female Marmosets - Converse - 2015 - The Journal of Sexual Medicine - Wiley Online Library

- Lululemon Unicorn Throwback Print Collection Is Back
:upscale()/2022/09/21/893/n/1922729/4b5f0e3b7b48f840_netimgAIwwvi.webp)
- Graduated double strand pearl necklace

- Women Underbust Waist Trainer Belt Slim Body Shaper Belly Wrap Compression Band

- Bodysuit Women Lingerie Fashion Lace S-3XL Underwire Jumpsuit Sexy Sleepwear Bra 34ddd Push up (Black, S) : : Clothing, Shoes & Accessories

- Plus Size - Retro Wired Swim Romper - Floral - Torrid
