Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction
By A Mystery Man Writer

Discovering molecular features of intrinsically disordered regions by using evolution for contrastive learning

Practical Applications of Computational Biology & Bioinformatics, 15th International Conference (PACBB 2021) [325, 1 ed.] 3030862577, 9783030862572

MECE: a method for enhancing the catalytic efficiency of glycoside hydrolase based on deep neural networks and molecular evolution - ScienceDirect

Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

Deep_CNN_LSTM_GO: Protein function prediction from amino-acid sequences - ScienceDirect

Sensors, Free Full-Text
IJMS, Free Full-Text
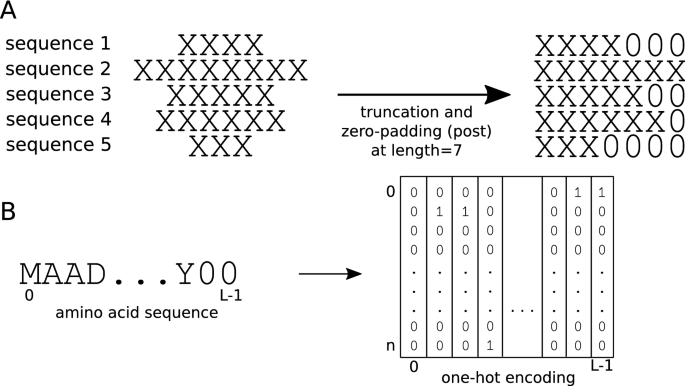
Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

PDF) Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

Materials, Free Full-Text

(PDF) Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

PDF] Effects of padding on LSTMs and CNNs

BoT-Net: a lightweight bag of tricks-based neural network for efficient LncRNA–miRNA interaction prediction

DeepRCI: predicting RNA-chromatin interactions via deep learning with multi-omics data
- box-sizing CSS-Tricks - CSS-Tricks

- Spacing Basics and Rules Every Designer Should Know

- Exploring Block Layout, Alignment, and Dimensions in WordPress

- What is the difference between the concepts of padding and width field in CSS? Both are working in the same way! - Quora

- Alignment, margin, and padding for layout - Windows apps
- Portrait Young Girl Underwear Stock Photo 246173359

- Buy DISOLVE� Women's Cotton Tummy Tucker Control Shaper or Slimming Panty Combo of 2Assorted Color Free Size Multicolour at

- Leggings femininas Savage X Fenty Special FX cintura alta média

- Brightech Embrace Led Spiral 2-in-1 Adjustable Floor & End Table

- Pink Diamond & White Beads Show Club Party Bra Gothic Soutien

